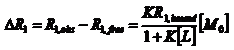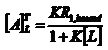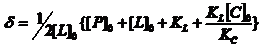|
Results: 2011 2012 2013 2014 2015 2016
2014
The strength of the ligand-macromolecule interaction, evaluated on the basis of the affinity index.
NMR has been widely used to study the interaction between small molecules and macromolecules. Among NMR methods employed for investigating ligand-macromolecules aducts, the evaluation of the so called “affinity index” is particularly efficient in order to compare the strength of the interaction process involving the same macromolecule and different ligands. If we consider the ligand –macromolecule equilibrium M+L↔ML with the equilibrium constant K=[ML]/[M][L] assuming [L]>>[M0], it has been shown that: where K is the thermodynamic equilibrium constant and [M0] is the initial macromolecule concentration. Since the ligand NMR parameter most affected by changes in the molecular dynamics is which was defined as “affinity index” [1]. The affinity index is a constant if temperature, T, and ligand concentration, L, are specified. In order to remove the effects of motional anisotropies and different proton densities a “normalized affinity index can be calculated as
Example:
[1] C. Rossi, A. Donati, C. Bonechi, G. Corbini, R. Rappuoli, E. Dreassi, P. Corti, Chem. Phys. Lett. 264, 205 (1997)
Results
Competitive binding experiments Competition binding experiments are well-established methods for determination of both ligand binding affinity and specificity to macromolecules. NMR based protocols employing competition methods expand the utility of existing experiments to characterize binding for compounds of higher affinity or those compounds that are sparingly soluble. a) We will consider the simple case in which two ligands L and C compete for the same binding site. Ligand L has a weak to medium binding affinity for macromolecule P and C has a high affinity for the same macromolecule. The monitored NMR parameter is
where
If the dissociation constant, KL, characterizing the binding of L, is determined from a previous experiment, the dissociation constant KC of the high affinity binder C, can be obtained. b) In another approach we consider the protein P with [P]0 =ct, a ligand L1 having a high affinity for P and a ligand L2 with a weak affinity for P. In the first step of the experiment we determine
where [P]free is the free protein concentration, after some fraction from [P]0 was complexed with L1 and is accessible for complexation with L2. We have [P]free = [P]0 – [PL1]. The mathematical expression of [PL1] can be obtained from the definition of the L1 dissociation constant KD(L1) = [P]free[L1]free/[PL1]. Solving a quadratic equation we obtain
[PL1]=
The expression [P]free = [P]0-[PL1] is substituted in the above equation and from the dependence of |

|
INCDTIM |